Searched for: hsa-mir-181d
hsa-mir-181d mutations modulate:
List of mutations that drive a causal flow containing a regulator followed by a downstream bicluster. Survival is the Cox proportional hazards regression of the bicluster eigengene versus paitent survival. Hallmark(s) is the hallmarks of cancer associated with the bicluster genes. Legend for hallmarks:
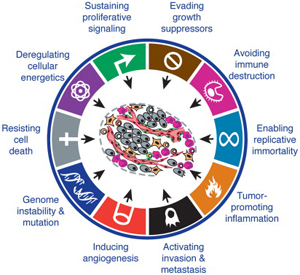
Hanahan and Weinberg, Cell 2011

Hanahan and Weinberg, Cell 2011
Mutations in hsa-mir-181d are not predicted modulate anything in gbmSYGNAL.
hsa-mir-181d regulates:
List of biclusters regulated by the regulator that was searched. Next to the regulator in parentheses is the regulators mechanism of action (Act. = activator and Rep. = repressor). Survival is the Cox proportional hazards regression of the bicluster eigengene versus paitent survival. Hallmark(s) is the hallmarks of cancer associated with the bicluster genes. Legend for hallmarks:

Hanahan and Weinberg, Cell 2011

Hanahan and Weinberg, Cell 2011
| Regulator | Regulated Biclusters |
|---|---|
| hsa-mir-181d | 15 |
| Regulator (Action) | Bicluster | Survival (p-value) | Hallmark(s) |
| hsa-mir-181d (Rep.) | PITA_438 | 3.48 (5.0E-04) | |
| hsa-mir-181d (Rep.) | PITA_76 | 3.19 (1.4E-03) | |
| hsa-mir-181d (Rep.) | PITA_122 | 2.57 (1.0E-02) | |
| hsa-mir-181d (Rep.) | PITA_516 | 2.62 (8.9E-03) | |
| hsa-mir-181d (Rep.) | TARGETSCAN_50 | 3.80 (1.4E-04) | |
| hsa-mir-181d (Rep.) | TARGETSCAN_151 | 2.60 (9.4E-03) | |
| hsa-mir-181d (Rep.) | TARGETSCAN_487 | 3.39 (6.9E-04) | |
| hsa-mir-181d (Rep.) | TARGETSCAN_533 | 4.20 (2.6E-05) | |
| hsa-mir-181d (Rep.) | TARGETSCAN_562 | 2.77 (5.6E-03) | |
| hsa-mir-181d (Rep.) | TARGETSCAN_592 | 3.17 (1.5E-03) | |
| hsa-mir-181d (Rep.) | TFBS_DB_51 | 2.51 (1.2E-02) | |
| hsa-mir-181d (Rep.) | TFBS_DB_125 | 3.99 (6.7E-05) | |
| hsa-mir-181d (Rep.) | TFBS_DB_428 | 2.62 (8.9E-03) | |
| hsa-mir-181d (Rep.) | PITA_262 | 3.22 (1.3E-03) | |
| hsa-mir-181d (Rep.) | PITA_262 | 3.22 (1.3E-03) | |
| hsa-mir-181d (Rep.) | TARGETSCAN_38 | 3.19 (1.4E-03) |
Biclusters containing hsa-mir-181d:
List of biclusters that contain the genes that was searched. Var. Exp. FPC is the variance explained by the first principal component and is a good measure of bicluster co-expression. Survival is the Cox proportional hazards regression of the bicluster eigengene versus paitent survival. Hallmark(s) is the hallmarks of cancer associated with the bicluster genes. Legend for hallmarks:

Hanahan and Weinberg, Cell 2011

Hanahan and Weinberg, Cell 2011
Not in any biclusters.